One of the most typical problems you face when you are trying to find periodicities in astronomical time series data is the sampling period. You may think (and if you’re a telecommunications engineer as I am, I bet you directly thought about it) about using the Fast Fourier Transform (FFT) or the DFT.
You can try that and you may obtain results that could be valid… or not. Especially if you are willing to analyze data in an iterative approach then you should not do this as you may end up facing a problem with some of your results (at least check the sample time in provided data).
Let’s take for instance this case: a file with magnitude time series data of a Cepheid star. As you may know you can use these stars as standard candles for some range of distances, Henrietta Leavitt realized in 1912 that there is a relationship between the periodicity of these stars and their luminosity. Therefore we can use this relationship to calculate the distance to a star. More about the method
here.
You can use any program you want to perform the FFT or DFT on the time series data, for instance Period04, which is a cool Java program that you may want to use for analyzing data (it could be helpful for some simple analysis about periodicities and could be used for prewhitening processes in asterosismology).
You can find Period04 program
here.
So, if you analyze this data using Period04 you will end up having this spectrum:
Where you can easily see that the main periodicity is located at 0.42 cycles/day (BTW: another note for engineers, as samples are taken on nearly
daily basis we will not be talking about Hz but c/d).
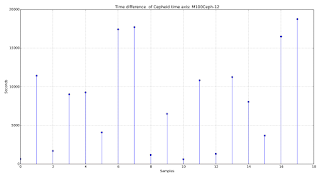
Well, you may think this is the way it is supposed to be… not so fast. Something I learnt while analyzing this type of data: mind the time axis, mate… The first tip you should extract from here is: don’t try to analyze something you don’t really understand: how was your data captured? What is it representing? And especially: is the sample time constant? If your background is electronics and DSP related, then you may be used to have a constant sample time. Forget about that. That may be true here but is not always true.
First thing first, let’s see what is the time difference between each sample. You can do it in matlab or python by using diff. If you are using python just import numpy (import numpy) and then use numpy.diff on your time data. These functions calculate something similar to the derivative, it just takes each sample and subtracts them one by one Therefore this will give us the difference between each sample. If you are not very familiar with Python then you will find an example about this at the end of this article. If you are used to code in Python then you will find this very very easy.
As we can see there is a huge difference in the sample time value. We should not use this particular method for analyzing this data. So, what can we use?
Well, there is a nice periodogram called the
Lomb-Scargle periodogram that will allow us to compute the spectrum of our time series but accepting these irregularities we saw in the sample time values.
This periodogram accept time lags in the basis functions it uses for projecting our data and searches for the best solution. We will be obtaining the most accurate spectrum by using this technique. Therefore forget about using FFT in this case and use this spectrogram, it could be slower but you will obtain a more accurate result.
Where Tau is the supported lag that will be used to make a least-square fit to maximize the orthogonality of both basis functions.
Ok then. Let’s use it!
You can find a nice implementation of this periodogram in Python
here.
The way of using it is (again: if you are not used to code in Python then you will find an example at the end of this article on how to use it and some data to test it) you should import the Lomb-Scargle periodogram before using it. If you placed the periodogram python code in the same directory then just do “import lomb” and we will be ready to go.
Now we should use this function call lomb.fasper(time_value, magnitudes_value, oversampling, Nyquist_freq)). Be aware that you should provide the function with the following parameters: times vector, magnitude values, the oversampling factor, and a factor that will be use to analyze up to Nyquist_freq*factor frequency.
The result is the following:
As you can see it is different. It turns out the that most important periodicity in this time series was not the one at 0.42 c/d but 0.047 c/d.
I hope this may help somebody trying to deal with this particular problem.
Take care!
Cepheids data: https://drive.google.com/file/d/0B3aZza3bav9gODBFR3VoQm04LWs/view?usp=sharing
PS: I prepared a cool Latex entry but it seems to have a huge amount of compatibility issues with blogger, I'm afraid format is quite poor. :-(
----------------------------
#!/usr/bin/python
import numpy as np
import matplotlib.pyplot as plt
import csv
import lomb
def proce(tipo):
names=list()
salida=list()
auxi=list()
for z in range(12):
aux=list()
if z+1<=9:
nombre="M100Ceph-"+'0'+str((z+1))+".txt"
else:
nombre="M100Ceph-"+str((z+1))+".txt"
names.append(nombre)
f=open(nombre,"rb")
k=csv.reader(f,delimiter="\t")
for row in k:
for z in row:
auxi.append(float(z))
aux.append(auxi)
auxi=list()
salida.append(np.array(aux))
domin=1 #Just change it if you want to see time domain signal
if tipo==1:
for k in range(len(salida)):
t=salida[k][:,0]
y=salida[k][:,1]
##---------
#Plotting time difference
plt.figure()
plt.stem(3600.0*np.diff(t))
plt.xlabel("Samples")
plt.ylabel("Seconds")
plt.title("Time difference of Cepheid time axis: "\
+names[k][0:len(names[k])-4])
plt.grid()
#---------
plt.figure()
if domin==1:
fs=(1/np.diff(salida[k][:,0]).max())
FOUR=salida[k][:,1]-(salida[k][:,1].mean())
Pun=len(y)
t,y, nout, jmax, prob = lomb.fasper(t,y, 6., 6.)
#print (prob)
print (salida[k][:,1].mean())
#y=y*y.conj()
plt.xlabel("Frequency (c/d)")
plt.ylabel("Magnitude")
plt.plot(t,y)
if domin==0:
plt.xlabel("Days")
plt.gca().invert_yaxis()
if domin==1:
plt.title("Lomb-Scargle periodogram of Cepheid: "\
+names[k][0:len(names[k])-4])
else:
plt.title(names[k][0:len(names[k])-4])
plt.grid()
plt.show()
return np.array(salida)
if __name__ == "__main__":
proce(1)